IRCM, Institut de Recherche en Cancérologie de Montpellier, INSERM U1194, Université de Montpellier ; and Institut régional du Cancer de Montpellier, Montpellier, F-34298, France
Under the effort of Prof. M. Ychou and Dr. AR Thierry a multi-disciplinary team was created taking advantage of the viscinity of the ICM (Institute of Cancer of Montpellier) and IRCM (Institute of Research on Cancerology of Montpellier) within the same campus to conduct translational research and to carry out fundamental research mainly related to a emerging diagnostic tool: the circulating DNA (cirDNA).
I. Joint IRCM/ICM effort: Creation of the IRCM team “Biomarkers for Precision Oncology”
A new team associating clinicians and researchers was created in 2014 under the leadership of Pr M. Ychou, in order to conduct integrative GI (Gastro-Intestinal) cancer research towards personalized and optimized medicine. This new team gathered diverse range of skills and expertises from several institutions all dedicated to basic and translational research in GI oncology and individualized therapy. The team entitled “Integrated Research in Digestive Oncology for Precision Medicine”.
The team first focused on discovering and developing early predictive and prognostic biomarkers for personalized management in GI oncology. Team’s plan is to establish new ties between fundamental research and clinical needs in order to achieve significant and innovative practice-advances in digestive cancer research.
The team has high visibility in the research and application of circulating DNA (cirDNA) as a diagnostic tool in oncology. A.R. Thierry is a key leader and recognized as a pioneer in this field. Based on several discoveries about the structure and origins of cirDNA, the team developed specific and optimal methods to analyze cirDNA allowing the first demonstration of the clinical validation and utility of cirDNA in oncology. Since January 2014, the team has completed 3 clinical assays and is engaged in nine other clinical trials, three of which have resulted in two ERC H2020 grants.
This program benefited greatly from the direction of Pr M. Ychou, Director of ICM and coordinator of the SIRIC grant, who is a recognized key leader in digestive oncology.
Dr A.R. Thierry co-leaded the team until 2020 and then leads the team entitled “Biomarkers for Precision Oncology”. Considering the expertise of the new team’s leader and the needs of the program development, the new team’s scientific strategy will focuse more on biomarkers research but will no longer be limited to digestive oncology and will extend to other cancer diseases.
In view of the high visibility of the team production on circulating DNA research, the team will now mainly focus on that field which is considered as one the most promising in the last decade in oncology together with the immune-therapy. Consequently, the team specific objectives will be to, first, continue to study and evaluate cirDNA in basic, technological and transational research; and second, to implement projects on biomarker discovery to potentially nourish new cirDNA applications.
II. Circulating DNA
Circulating cell-free nucleic acids (cfNA) is one of the fastest growing and most exciting areas in oncology in recent years. Circulating cell-free DNA/RNA (cirDNA/RNA) is defined as extra-cellular DNA/RNA occurring in blood. Several observations, including ours, suggest that the study of these NAs, especially cirDNA, has a great potential, especially for cancer screening, prognosis and monitoring of the efficacy of anticancer therapies.
As liquid biopsy, cirDNA collected from blood, have many advantages over tissue biopsies:
- Plasma samples are compatible with genetic analysis. Genetic analysis from tissue biopsies face challenges in mostly using preserving reagents potentially damaging DNA. In contrast, the preservatives used in blood samples do not negatively affect current genetic analysis methods.
- Collection is less invasive: CirDNA is easily extracted from blood samples, and blood draw is minimally invasive as compared with surgical resection or needle biopsy, and as a result, lowering the risk to patients.
- Heterogeneity is detectable: liquid biopsies contain information about all parts of a tumour in patients and from multiple tumour sites in patients with multiple metastases. This wider genetic picture contributes to a more accurate characterization of the molecular profile, enabling therapy guidance.
- Samples include information about early metastatic lesions: tissue biopsies are performed only on tumours that have already been detected, by imaging or other analytical methods. Liquid biopsies include additionnal information about minimal residual disease and metastatic lesions that may not yet have been detected, enabling clinicians to identify patients in the need of treatment.
Because of the strong clinical need about theragnostics/drug predictive information, especially with the advent of the targeted therapy, detection of actionable mutations to guide clinician towards appropriate and personalized therapy, was historically the first cirDNA clinical objective in oncology. However, cirDNA potential should not be restrained as only a “liquid biopsy” but also as a powerful source of biomarkers and, as such, as a useful tool for patient follow up. Since blood samples can be collected at multiple time points during therapy and follow up, it shows higher advantages than invasive procedures. Thus, blood samples can be more easily and safely obtained than tissue biopsies during the course of therapy and follow up. Blood samples can be drawn regularly at routine clinical visits and do not require highly trained surgical personnel or specific equipment. This allows for dynamic monitoring of molecular changes in the tumour over the course of treatment rather than relying on a static time point at the initiation of treatment.
III. Contribution to the cirDNA research and applications
III.1 Team’s objectives
Following their first clinical evaluation in predictive medicine, the potential of cirDNA analysis now covers each phase of cancer patient management care by examining the predictive information, the detection of the minimal residual disease, the early detection of resistance, treatment monitoring, recurrence surveillance, and cancer early detection/screening. However, the lack of standardization of the pre-analytical conditions hinders the use of cirDNA as a clinically robust biomarker in oncology. Lastly, features and potential biological functions of cfNA are poorly known. For instance, increased knowledge in specific cirDNA structure, as observed by size profile analysis, methylation or nucleosome positioning may benefit to higher capacity in diagnostics or cancer screening.
III.2 Pioneering findings and scientific achievements
We started to work in 2005 on circulating DNA despite no financial support up to 2010 since private and public agencies denied interest in this area; for instance, the refusal notice of two independent funding agencies we received at that time, indicated that cirDNA was “science fiction”.
Structure and origins of cirDNA
We initially provided pioneering observations on the structure and origins of tumor-derived ccirDNA.
1-We designed an experimental mouse model for studying ctDNA content variation with tumor progression. We found that plasma rather serum is the appropriate biological source. We determined the origin and performed quantification of circulating DNA in mice with human colorectal cancer xenografts (Nucleic Acid research, 2010).
2-We discovered that high fragmentation characterizes tumor-derived circulating DNA (PlosOne, 2011).
3-We pointed out that the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients is of particular importance (Expert Opinion Mol Biol, 2012)
4-We reported a higher cirDNA analytical detection capacity when analyzing single strand DNA (NPJ Genomic Medicine, 2018).
5-We characterized the mitochondrial cirDNA release in healthy and cancer individuals (Sci.Rep. 2019).
6-We demonstrated the difference in cirDNA size fragment analysis (termed Fragmentomics) when comparing plasma of cancer and healthy individuals using qPCR, and Whole Genome Sequencing (WGS) from single or double strand DNA library preparations (NPJ Genomic Medicine, 2018).
7-We elucidated the structural forms of cirDNA nucleo-proteic complex in the blood stream (NPJ Genomic Medicine, 2018; Nucleic Acid Res, 2019).
8-We evidenced the influence of hypoxia on the cirDNA release from cancer cells in vitro and in vivo (British J Cancer, 2019).
9-We demonstrated the presence of cell-free intact mitochondria circulating in blood (FASEB J., 2020).
10-We first revealed the full fragment size distribution of nuclear and of mitochondrial cirDNA (JCI Insight, 2021; J.Biol.Chem. 2022)
Technological studies
1. We validated the multimarker approach of IntPlex. IntPlex is the first and so far unique technology enabling qualitative and quantitative multiplex analysis for ctDNA (Mol.Oncol., 2013)
2. We provided the first initial guidelines for the pre-analytical conditions for ctDNA analysis. (Clin.Chem.Acta, 2013)
3. We provided the first full guidelines on the pre-analytical conditions for cirDNA analysis (Clin.Chem. 2019).
4. We provided the first full guidelines on the pre-analytical conditions for cirDNA methylation analysis (Clin.Epigenetics, 2021).
5. We demonstrated a higher cirDNA analytical detection capacity when examining single strand cirDNA size profile with shallow Whole Genome Sequencing (sWGS) (Npj Genomic Medicine, 2018).
6. We demonstrated that fragmentomics as determined by sWGS and qPCR concurrent analysis showed strong potential in discriminating plasma from mCRC to healthy patients (JCI Insight, 2021.
7. By combining the MNR test, quantitative analysis and fragmentomics and assistance of machine learning (Artificial Intelligence), high level of performance in screening cancer individuals was obtained in a study involving more than 1200 individuals (Adv.Sci, 2020).
8. We have registered patents on the use of cirDNA for survival prognosis, biomarker and screening test on the use of circulating mitochondrial DNA, cancer screening test by examining cirDNA size profile with shallow Whole Genome Sequencing (sWGS) from single strand DNA library preparation and from double strand DNA library preparation; and on a q-PCR based test for detecting the EGFR mutations.
Translational studies
1. We highlighted that cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. (Transl.Oncol. 2013)
2. We demonstrated that mutant cirDNA fragments are shorter than wild type cirDNA fragments in mCRC patient plasma (Transl.Oncol. 2013).
3. We were the first to clinically validate (in a prospective, blinded, large cohort and multicentric study) the analysis of ctDNA in oncology with a clinical prospective, blinded, multicentric study: Clinical validation of cirDNA analysis when detecting KRAS and BRAF mutations in metastatic colorectal cancer patients. (Nature Med. 2014).
4. We were the first to demonstrate (in a prospective, blinded, large cohort and multicentric study) the prognostic value of the cirDNA analysis in oncology: Circulating DNA as a Strong Multimarker Prognostic Tool for Metastatic Colorectal Cancer Patient Management Care (Clin.Cancer Res. 2015).
5. We detected the emergence of RAS and BRAF mutations following anti-EGFR therapy in mCRC patients and showed the convergent evolution and common resistance mechanisms during Treatment of Colorectal Cancer (Clin.Cancer Res. 2016).
6. We were the first to demonstrate (in a prospective, blinded, large cohort and multicentric study) the clinical utility of cirDNA analysis in oncology: Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment (Ann.Oncol., 2017))
7. CirDNA as a follow up parameter to monitor Sorafenib activity in mCRC patients (Mol.Oncol. 2021).
8. Neutrophil extracellular traps (NETs) production is associated with metastatic colorectal cancer (iScience, 2022) and COVID-19 (J.Clin.Med., 2020, Clin.Cancer Res. 2021)
IV Projects and strategies
IV.1 Work plan
Basic research
Broadening our knowledge of the structural and biological features of cirDNA should provide new avenues of research in tumor biology and help in standardizing the pre-analytical conditions, which remain key steps to be taken towards the clinical implementation of cirDNA analysis. Objectives are to:
1. Decipher the structural features of circulating DNA (cirDNA) with using various and complementary methods of WGS and of Q-PCR.
2. Study the mitochondrial cirDNA release, structural forms and relation to disease stages and particular physiologic conditions (with inflammatory process, RO species, hypoxia or ischemia…).
3. Determine the respective proportions of the various cirDNA structural forms and nucleo-proteic complex.
4. Determine cirDNA peri-surgery kinetics in stage II-III cancer patients (collaboration with CHU Nimes).
5. Investigate/develop circulating messenger RNAs (cf-mRNAs) as cancer biomarkers,
Technological research
1. Implement novel analytical parameters for optimizing cfNA analysis.
2. Evaluate mitochondrial genes sequences as biomarkers for analyzing cirDNA.
3. Standardize cirDNA analysis
4. Set up a single strand DNA library preparation toward routine clinical use to analyse cirDNA (collaboration with Integragen, Paris, France)
5. Determine cirDNA fragmentation parametrics in healthy and cancer subjects (MiTest project, MSD Avenir)
Translational research
Assess further clinical unmet needs by examining in clinical studies: the detection of the minimal residual disease, the early detection of resistance, treatment monitoring, recurrence surveillance, and cancer early detection/screening. Objectives are:
1. Detection of the minimal residual disease and surveillance of the recurrence in stage II-III CRC postsurgery patients (DNAcirc, INCA PRTK). Ongoing and ending in 2021. N= 470 patients; Prospective and multicentric study. AR Thierry, co-coordinator.
2. First interventional study in using cirDNA as test for selecting patient for anti-EGFR therapy in mCRC patients (Panirinox, AMGEN). Unicancer, promotor. N= 480 screened patients. Prospective, open-label and multicentric study T. Mazard, coordinator; AR Thierry, Scientific coordinator.
3. Comparison of cirDNA and radiological analysis in locally advanced breast and rectal cancer (LIMA, ERC H2020 EU). N= 220. Prospective, blinded and multicentric study (Utrecht University Hospital, ICM). AR Thierry, partner.
4. Evaluation and evolution of a personalized molecular tag in post-surgery stage III CRC patient (THRuST, ERC H2020 ERA-NET). N=320. Prospective, blinded and multicentric study (ICO, Barcelona; CIS, Torino; VHIO, Barcelona;) AR Thierry grant coordinator.
5. CirDNA as a follow up parameter to monitor Sorafenib activity in mCRC patients (Texcan, Gercor/Bayer).
6. CirDNA analysis in the course of pancreatic cancer treatment (Gabrinox assay, E. Assenat, CHU Montpellier)
7. Breast cancer prognosis by circulating tumor DNA analysis with using multiparametric structural parameters test (within MyProbe project, IGR, F. André). N=300. Retrospective, blinded and multicentric study. AR Thierry, partner.
8. Lung cancer prognosis by cirDNA analysis with using multiparametric agnostic parameters test (within the REVEAL project, IGR, B. Besse). N=1600. Prospective, blinded and multicentric study. AR Thierry, partner.
9. Evaluation of a multiparametric test for cancer screening in various cancers (MiTest, MSD MiTest and DepLR studies). N=1700. AR Thierry, coordinator.
10. Diagnosis and follow up of COVID-19 and cancer patients by NETs production analysis from various ancilliary studies (N~900).
IV. 2 Expected results and impacts in oncology
Deciphering the structural features of cirDNA upon disease stages and determining their respective proportion should directly improve the detection sensitivity of cirDNA and specifically improve their performance as diagnostic markers. In light of this, the study of the mitochondrial cirDNA release, structural forms and relation to disease stages and particular physiologic conditions would provide clues on links of cirDNA release and tumor progression with particular physiologic conditions. In addition, mitochondrial cirDNA may bear in some cases genetic alterations that would be of diagnostic interest. CirDNA half-life has been very poorly investigated although such knowledge is of obvious interest in regards to better collecting blood and about pre-analytical conditions, especially in respect to use of longitudinal analysis. Study the relationship between cirDNA release and NetoSis/DNA traps would provide clues in regards to metastasis formation and inflammatory process and immune defense.
We are convinced that the clinical trials will have a significant impact on the development of cirDNA in terms of clinical validation and utility as a novel diagnostic biological source towards more efficient cancer patient management care. For instance:
– The detection of the Minimal Residual Disease might help the oncologist to better select patients eligible for adjuvant therapy after surgery in stage I-III patients.
– The study of surveillance of the recurrence would be the first study to delineate the diagnostic and prognostic role of cirDNA in stage I to III CRC patients. Positive outcome of this study would subsequently enable to evaluate the clinical utility of the cirDNA as a new diagnostic approach to improve the patient surveillance tool arsenal. The surveillance of cirDNA for patients able to support a curative treatment should improve detection of relapses and decrease false positive and cost of the surveillance. This strategy could allow early systemic therapy and improve overall survival.
– We will propose the first interventional plasma DNA analysis for the detection of RAS/BRAF hotspot mutations in a clinical trial on anti-EGFR therapies.
– The prospective study of the emergence of RAS/BRAF/EGFR/PIK3CA mutations in mCRC patients undergoing anti-EGFR therapy. The diagnostic technology developed by the project will also improve prognoses by providing clinicians with information about the biological and genetic characteristics of the disease in each individual patient over time, allowing them to prescribe the right therapy at the right time. Impacts would be: the validation of a strategy for earlier diagnosis, patient stratification and better prognosis, improved clinical decisions and better health outcomes more sustainable health care systems.
– Since prevention or early diagnosis are the most efficient means to fight cancer, it is worthwhile to evaluate a screening method based upon cirDNA for cancer patients. We are aware that we are in the prehistorical stage of developing of such a test but it is meaningful to build the fundations of such a challenge despite it requires important ressources.
V. Discovery to translational research
Biomarker research and development may be constituted by three themes respective to markers: Mechanistic, therapeutic, and clinical disease:
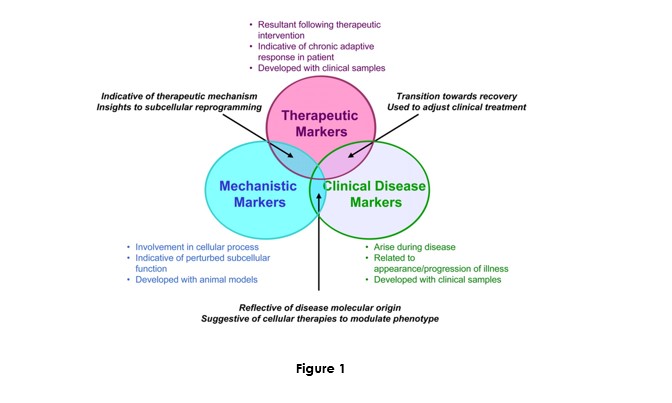
The originality of our research is located at the intersection of these three themes. It will foster research on circulating nucleic acid as a new diagnostic biological source and applying them with known or new genetic and epigenetic markers.
This strategy is only possible when uniting a multidisciplinary team with principal investigators with needed skills: molecular biology (P. Blache, AR Thierry), cellular biology and animal models (C. Prevostel), clinical biomarker (E. Crapez), biostatistics (C. Mollevi), and oncology (M. Ychou, A. Adenis, T. Mazard and B. Roch). The relationships with national and international networks in each domain especially in regards to the two European consortium and involvement in two RHU programs, enlarge the scope of knowledge/skills and vision of the biomarker field in concology. We are convinced that our strategy would contribute to successful research towards implementation of improved biomarkers in precision oncology.
VI. ICM efforts
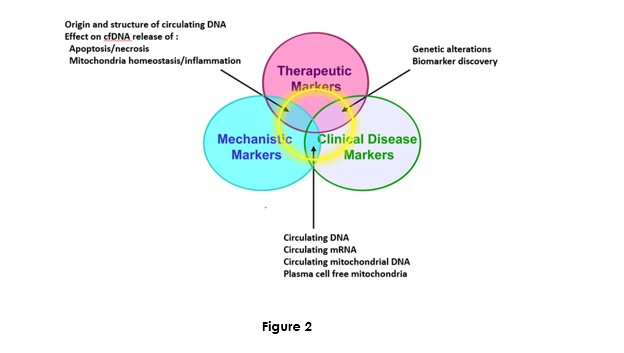
Dr M. Ychou Director of the ICM and of the SIRIC network Grant is fully convinced on the potential of cirDNA in oncology. In addition to its input to the IRCM team, and in contributing and designing translational research programs, he favored building plasma banking of plasma to facilitate actual research programs and anticipate the future needs, especially in extending the scope of the research on various cancer types (Figure 2). Most of the clinical studies sponsored by the ICM proposed a plasma banking for anticipating the use of cirDNA analysis. The ICM has the objective to be a leading clinical center about the translational research on cirDNA.
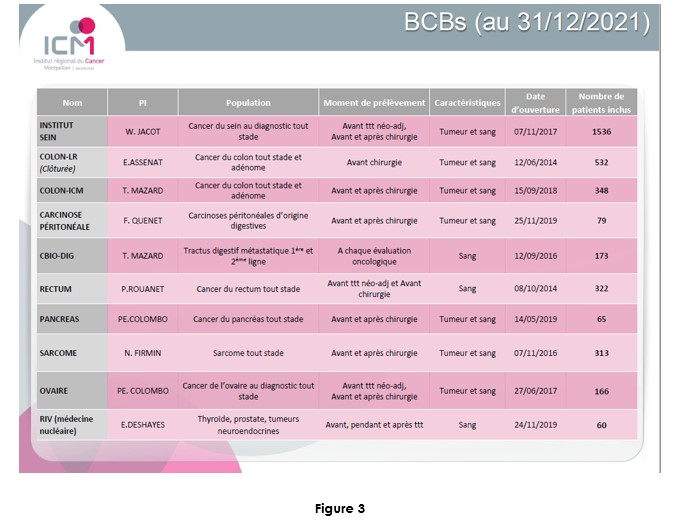
Conclusion
The cirDNA field is now popular in oncology and subject to exponential reporting in the literature. For instance, prescription in oncological theragnostic is allowed in several clinical conditions (lung cancer, melanoma and mCRC for guiding the clinicians about targeted therapy). Owing to our work and to the international efforts from leading teams (Among others: N. Rosenfeld, Cambridge, UK; L. Diaz, New York, USA; L. Bardelli, Torino, Italy; N. Turner, Manchester, UK; S. Kopetz, Houston, USA; S. Holdenrieder, Munchen, Germany; M. Diehn, San Francisco, USA; C. Andersen, Aarhus, Denmark; and P. Laurent-Puig P and V. Taly, Paris; …) other cirDNA potentials such as the detection of the Minimal Residual Disease or therapy efficacy monitoring, should extend their diagnostic applications in cancer management care. Furthermore, considering the intense cirDNA-related research now being carried out, especially with regard to cirDNA methylation or fragmentomics and assistance of Artificial Intelligence, we are convinced that the scientific community might be able to obtain in the next decade the oncology’s ‘grail’: cancer screening.
Acknowledgements
We thank the IRCM team members, T. Mazard, A. Adenis, B. Roch, M. Ychou, P. Blache, C. Prevostel, E. Crapez, F. Frayssinoux, L. Lasorsa, E. Pisareva, B. Pastor, C. Sanchez, A. Kudriatsev, A. Mirandola, M. Grosgeorge; IRCM Director C. Sardet; I. Moussion and S. Thezenas (ICM DRCI); S. Thezenas; V. Guillaumon and K. Saget (SIRIC Montpellier). AR Thierry is supported by the INSERM. This work was partially granted from the SIRIC Montpellier Cancer Grant INCa_Inserm_DGOS_12553.










